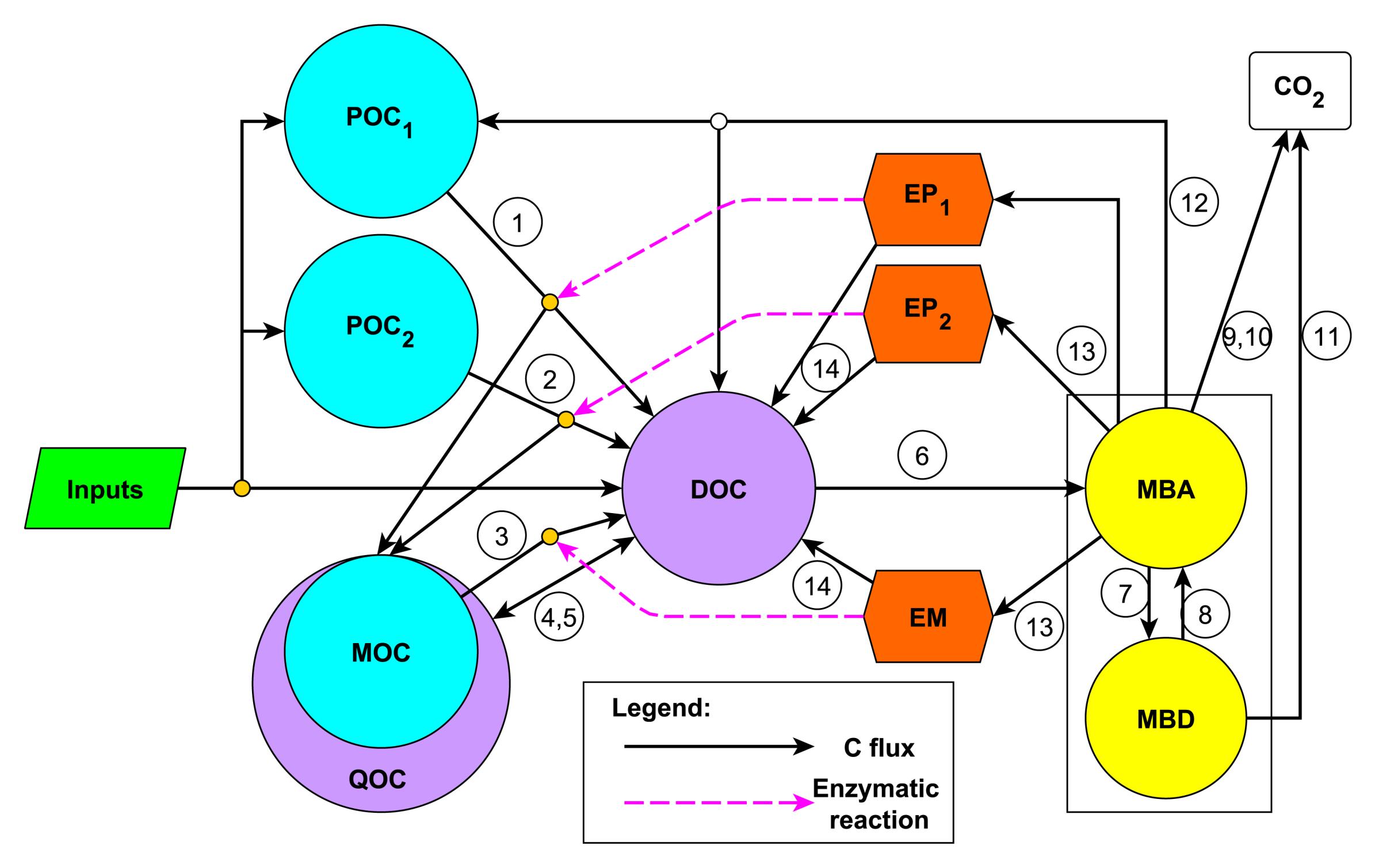
Microbial ENzyme Decomposition model (MEND)
We developed MEND because we observed that most Earth System Models lacked mechanistic details about microbial decomposition, including adsorption and desorption of dissolved organic carbon, active microbial biomass, and enzymes. Further most Earth System Models use conceptual rather than measurable soil pools and first order decomposition rates.
Our model has three soil carbon pools:
- particulate organic carbon as cellulose (POC1),
- particulate organic carbon as lignin (POC2), and
- mineral associated organic carbon (MOC).
With enzymes EP1, EP2, and EM corresponding to the soil pools.
Enzymes decompose the carbon pools via the Michaelis-Menten equation, resulting in the production of dissolved organic carbon (DOC) that can be taken up by microbes (MBA and MBD).
Testing and Validating the Model
We initiated a set of long-term incubation experiments to test and validate the model, involving four 14C labeled substrates and four soils, and using bulk soils and fractions of mineral-associated and particulate organic carbon. We found that the predicted microbial biomass was unreasonably low over long time frames, which suggested that large portions of the microbial community became dormant over time in the incubations. We developed a physiological model to account for active (MBA) and dormant (MBD) microbial fractions in MEND, and a new testing method for incubations over short and long time frames. New incubation experiments are ongoing.
The model is also being tested in the field, with the goal of experimental separation of contributions of mycorrhizae, bacteria, and root respiration, which will lead to additional model revisions. In addition, MEND is being incorporated into the Accelerated Climate Model for Energy (ACME), DOE’s next-generation Earth System Model.
Publications and Features
Feature article in Science News for Students, Recycling the dead: The decay of dead things is simply nature’s way of mining old materials for new uses. Kathiann Kowalski (2014) https://student.societyforscience.org/article/recycling-dead
Wang, G., Jagadamma, S., Mayes, M.A., Schadt, C., Steinweg, J.M., Gu, L., and Post, W.M. 2014. Microbial dormancy improves development and experimental validation of ecosystem model. The ISME Journal doi:10.1038/ismej.2014.120.
Jagadamma, S., Steinweg, M., and Mayes, M.A. 2014. Influence of substrate chemistry on carbon decomposition and microbial community composition. Biogeosciences 11:4665-4678.
Li, J., Wang, G., Allison, S.D., Mayes, M.A., and Luo, Y. 2014. Soil carbon sensitivity to temperature, carbon use efficiency, and model complexity in two microbial-ecosystem models. Biogeochemistry doi:10.1007/s10533-013-9948-8.
Jagadamma, S., Steinweg, J.M., Mayes, M.A, Wang, G., and Post, W.M. 2013. Mineral control on decomposition of added and native organic carbon in soils from diverse eco-regions. Biology and Fertility of Soils 49, doi: 10.1007/s00374-013-0879-2.
Wang, G., Post, W.M., Mayes, M.A. 2013. Parameterizing an enzyme-mediated soil organic carbon decomposition model. Ecological Applications, 23(1): 255-272. doi: 10.1890/12-0681.1.
Wang, G., Post, W.M., Mayes, M.A., Frerichs, J., and Jagadamma, S. 2012. Parameter estimation for models of ligninolytic and cellulolytic enzyme kinetics. Soil Biology & Biochemistry 48:28-38, doi 10.1016/j.soilbio.2012.01.011.
Data Products
Jagadamma, S., Mayes, M.A., Steinweg, J.M., Wang, G., Post, W.M. 2014. Organic Carbon Sorption and Decomposition in Selected Global Soils. Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, U.S. Department of Energy, Oak Ridge, Tennessee, U.S.A. http://dx.doi.org/10.3334/CDIAC/ornlsfa.002 .